4. Graph Data
"Lost my shape - trying to act casual! Can't stop - I might end up in the hospital I'm changing my shape - I feel like an accident They're back! - to explain their experience" - Talking Heads "Crosseyed And Painless" [Album: Remain in Light]
Introduction
This chapter covers the conversion of the catalog data to RDF triples, data validation using SHACL, and upload to the database.
Data model
"Whatever data you manipulate or store, there is always a model (also referred to as "schema" or "ontology"). Sometimes this model is not explicit, but it exists nevertheless, at least in the developers heads and is consequently reflected in the code that is written to manipulate or consume graph data." - Sequeda and Lassila (2021)
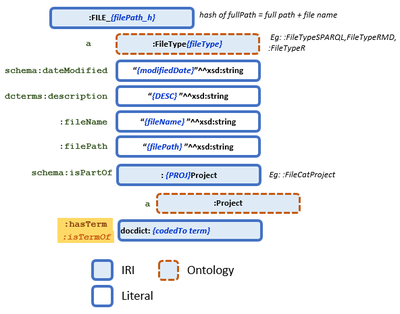
Before converting the data to RDF, it is useful to create a diagram of the data model based on the draft ontology, competency questions, and data extracted from the files. The model evolves during code development, improving with each iteration. The flexibility of the graph data facilitates this flexible approach to model development.
A diagram like Figure 4.1 helps verify your thinking during the development process. It is also helpful later when writing SPARQL queries.
Data Conversion
There are many approaches and tools that can be used to convert the collected data to RDF triples. For example, mapping languages like R2RML are used to map relational sources to graph databases. This project uses R code for the data conversion, providing a high level of control of the data and structure at the expense of writing and maintaining many lines of R script.
The R code below illustrates a for loop that processes the dataframe, first assigning a FileType class to the file IRI. Next, if a value for file modification date is present in the data, a triple is formed to represent this information. Many more lines of code will follow to fill out the data model with instance data.
for(i in 1:nrow(srcDf))
{
# File Type
rdf_add(some_rdf,
subject = paste0(FILECAT, "FILE_", srcDf[i,"filePath_h"]),
predicate = paste0(RDF, "type"),
object = paste0(FILECAT, "FileType", srcDf[i,"fileType"])
)
# Date Modified
if ( ! is.na (srcDf[i,"modifiedDate"]) ) {
rdf_add(some_rdf,
subject = paste0(FILECAT, "FILE_", srcDf[[i,"filePath_h"]]),
predicate = paste0(SCHEMA, "dateModified"),
object = srcDf[i,"modifiedDate"],
objectType = "literal",
datatype_uri = paste0(XSD,"dateTime")
)
}
.... more lines of code
For example, an R file type with a date last modified of 2021-08-18T13:34:31 is represented with instance data similar to:
FILE_03c754f5 rdf:type :FileTypeR
FILE_03c754f5 schema:dateModified “2021-08-18T13:34:31”^^xsd:dateTime
The file Subject IRI is created using a hash of the full path to the file, ensuring a unique identifier for that entity. The hash is truncated to eight characters for readability. Strategies for IRI creation and resolution are beyond the scope for this documentation.
After the data file is created it is validated using SHACL.